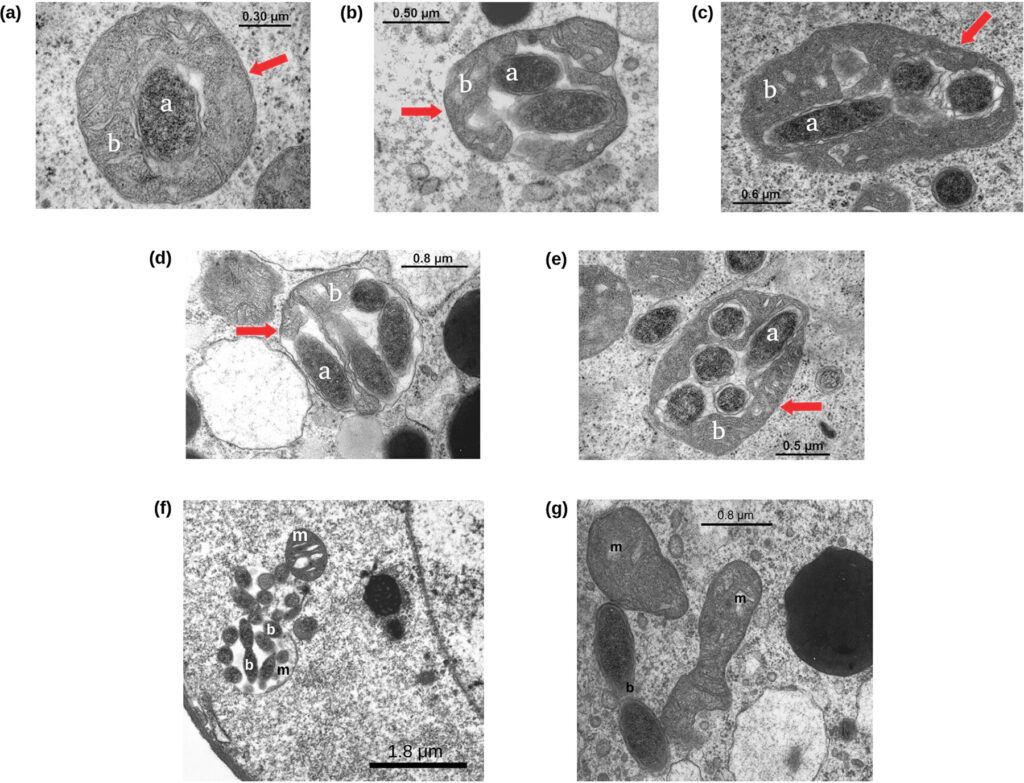
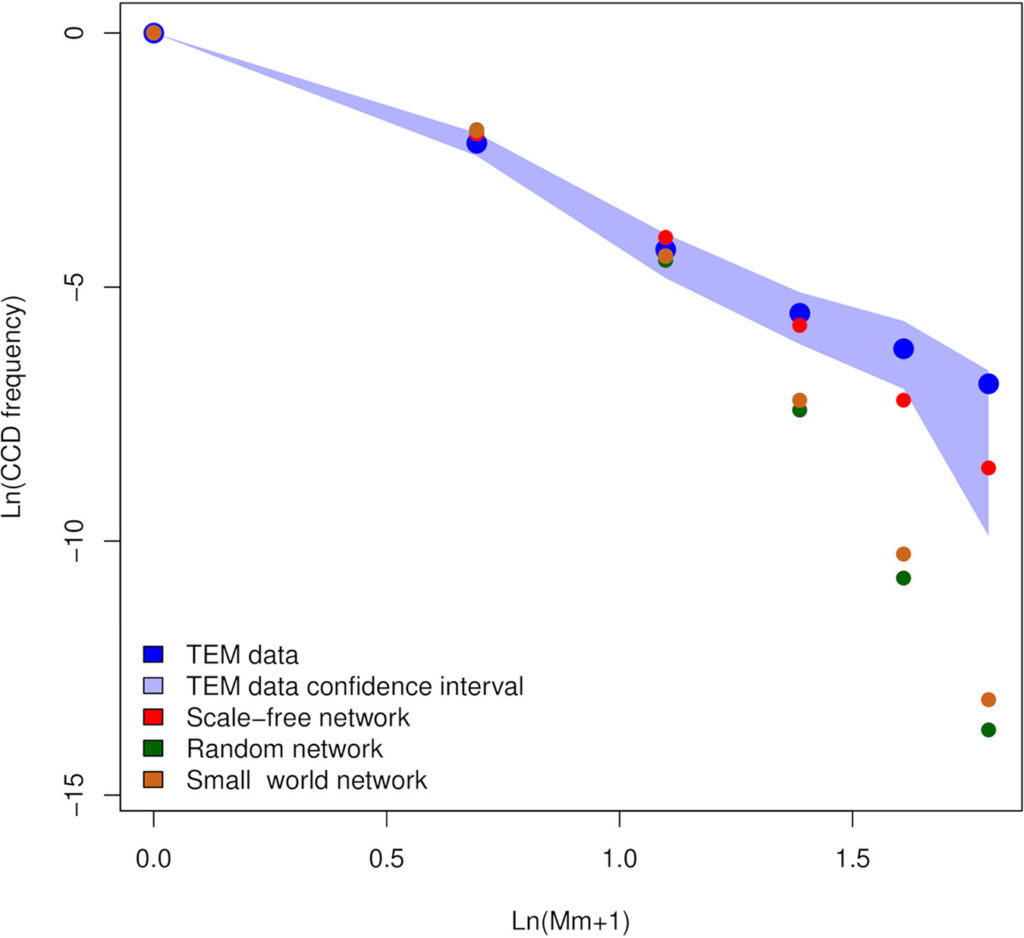
Summary of: Sterzi, L., Nodari, R., Di Marco, F. et al. Genetic barriers more than environmental associations explain Serratia marcescens population structure. Commun Biol 7, 468 (2024). https://doi.org/10.1038/s42003-024-06069-w
In bacterial species we often observe a remarkable genetic diversity, with the presence of clear distinct genetic clusters. The view on how these genetic clusters are formed and maintained (i.e. speciation models) initially focused mostly on the interaction between mutations, selection and ecological divergence. These forces create a dynamic equilibrium where new lineages continuously emerge by mutation and compete for the same ecological niche, until one eventually out-competes the others. In this view, diversity emerges mainly when a mutation allows it to occupy a novel ecological niche: each lineage represents an “ecotype”. In the last decades, the dynamic nature of the bacterial gene repertoires and the extent of recombination (i.e. gene flow and horizontal gene transfer) in bacterial evolution have led to suggest a speciation model where genetic diversity is enhanced and maintained by barriers to recombination between subpopulations. In this study, we investigated how ecological divergence and genetic barriers could explain the population structure of Serratia marcescens, an opportunistic pathogen with high ecological and genetic plasticity.
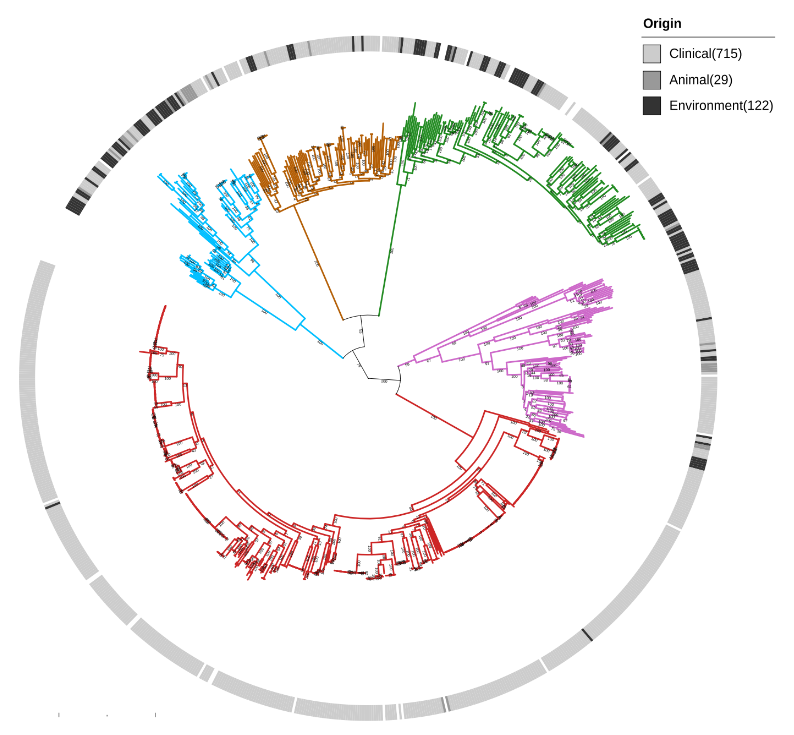
We show that Serratia marcescens comprises five deeply-demarcated genetic clusters. Analysing gene content and isolation sources, we observed that only two clusters show consistent signals of ecological specialisation, with one cluster strongly associated with clinical settings. We also used cluster-specific genes to investigate the occurrence of the genetic clusters in bacterial communities sampled from different biomes. We noted that the clusters co-occur in several biomes, but we also highlight that the clinically-associated cluster is enriched in freshwater samples, suggesting a possible reservoir.
We studied gene flow within the species and, interestingly, we observed that genetic exchange between the five clusters in Serratia marcescens is notably limited. For this reason, we focused our attention on Restriction-Modification (R-M) systems. These systems are widespread bacterial defence systems which remove exogenous DNA, relying on a methyltransferase which methylates a specific sequence motif on the endogenous DNA and on a cognate restriction endonuclease which cleaves DNA when the motif is unmethylated. We observed a partial incompatibility of R-M systems coherent with limited inter-cluster gene flow, and we also noted that clusters are enriched in different types of R-M systems.
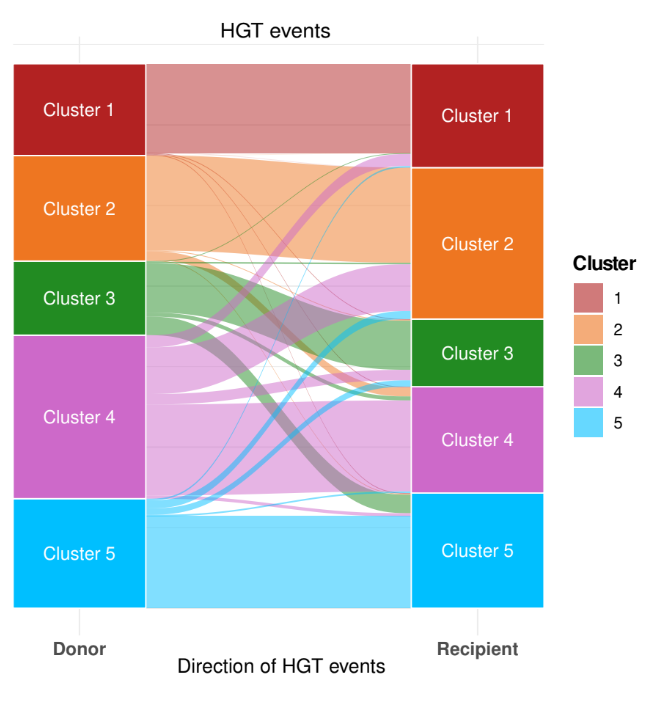
In conclusion, we believe that genetic barriers play a large part in maintaining diversity in subpopulation of S. marcescens. Within this population structure, two clusters have initiated adaptive trajectories to specific ecological niches and proceed to progressively isolate from the others. Whereas, other clusters are ecologically generalist and despite they often co-occur in the same environment at the same time, genetic barriers are sufficiently thick to maintain the clusters regardless of ecology or spatial distribution. Thus, with a hint of speculation, we propose that the leading role in the evolution of S. marcescens is played by the genetic barriers between co-occurring, ecologically generalist subpopulations.